
An on-line database of predicted and experimentally determined protein-protein interactions.

An on-line database of predicted and experimentally determined protein-protein interactions.

A database of RNA interactome from sequencing experiments.
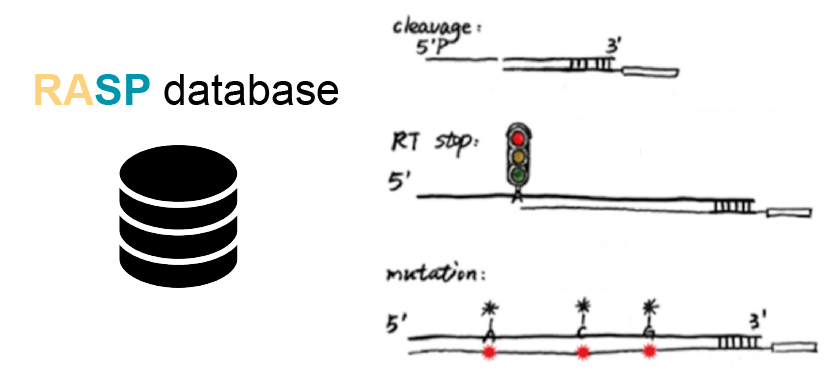
An atlas of transcriptome-wide RNA secondary structure probing data
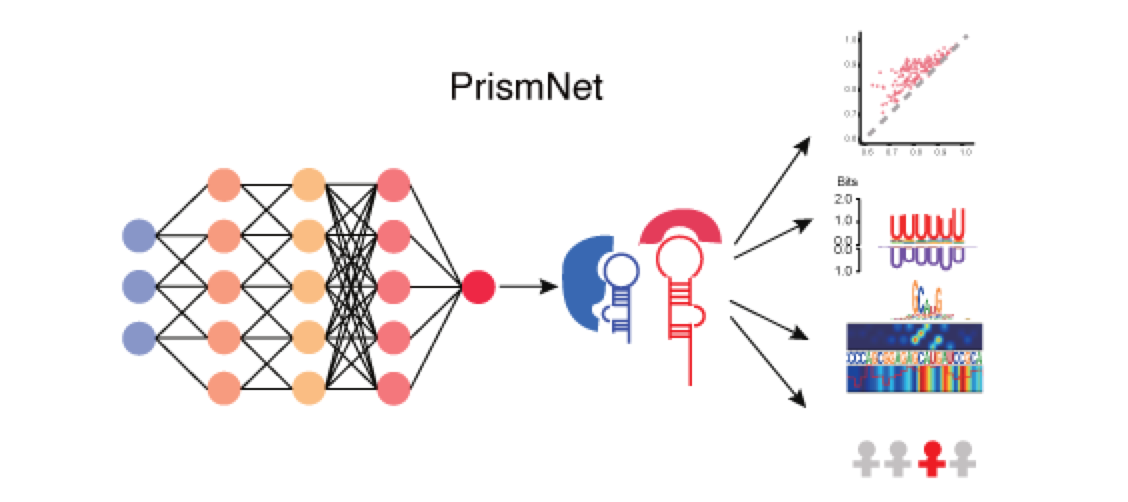
A deep learning tool that integrates experimental in vivo RNA structure data and RBP binding data for predicting dynamic cellular protein-RNA interactions
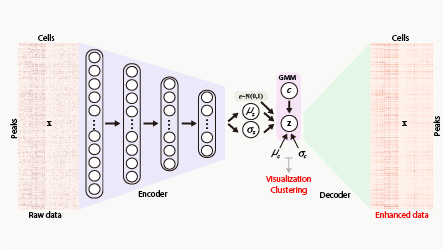
A deep learning tool for single-cell ATAC-seq analysis including visualization, clustering and imputation
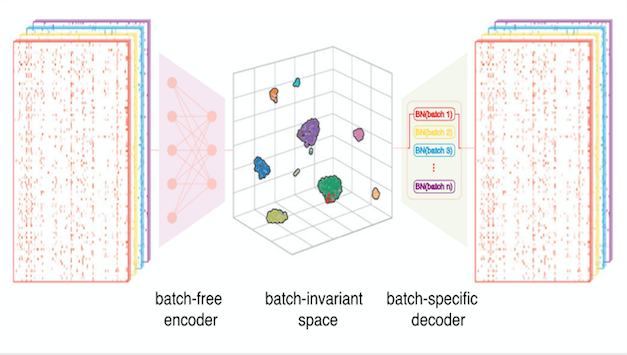
A deep learning tool for single-cell sequencing data integration
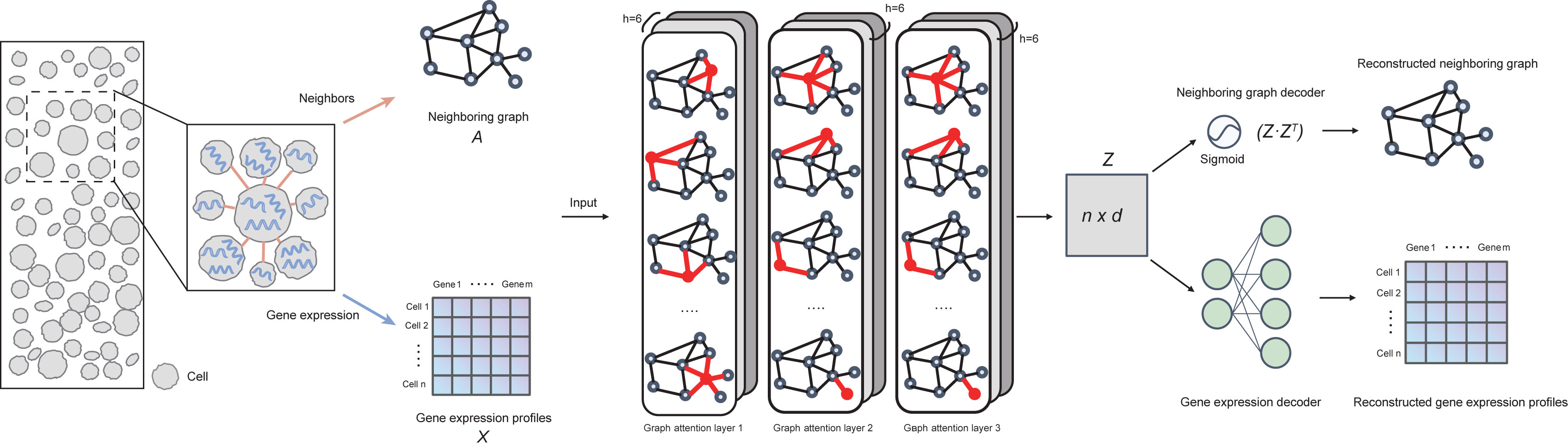
A deep learning tool for spatial transcriptomics analysis via cell embedding

Data Link: GSE64169
Paper Link: Structural imprints in vivo decode RNA regulatory mechanisms