rRNA
Human and mouse rRNA copies are collected from SILVA. And precursor rRNA from NCBI is collected. The matured rRNA is the spliced from pre-rRNA. Below is structure of pre-rRNA.
5_spacer(5'ETS) 18S spacer_1(ITS1) 5.8S spacer_2(ITS2) 28S 3_spacer(3'ETS) IGS SP IGS UCE/CPE
<================><=====><================><=====><================><=====><================><=========================><======><========><=========>
rRNA sequences are located in /150T/zhangqf/GenomeAnnotation/rRNA
tRNA
Human and mouse tRNAs are collected from Gencode and GtRNAdb. GtRNAdb contains tRNA gene predictions made by tRNAscan-SE on complete or nearly complete genomes. Unless otherwise noted, all annotation is automated, and has not been inspected for agreement with published literature. Predicted secondary structures are also provided.
tRNA sequences and structures are located in /150T/zhangqf/GenomeAnnotation/tRNA
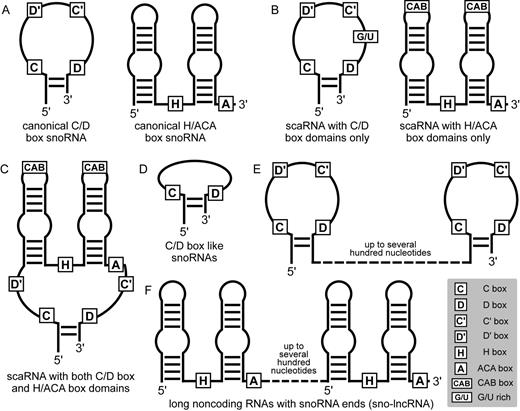
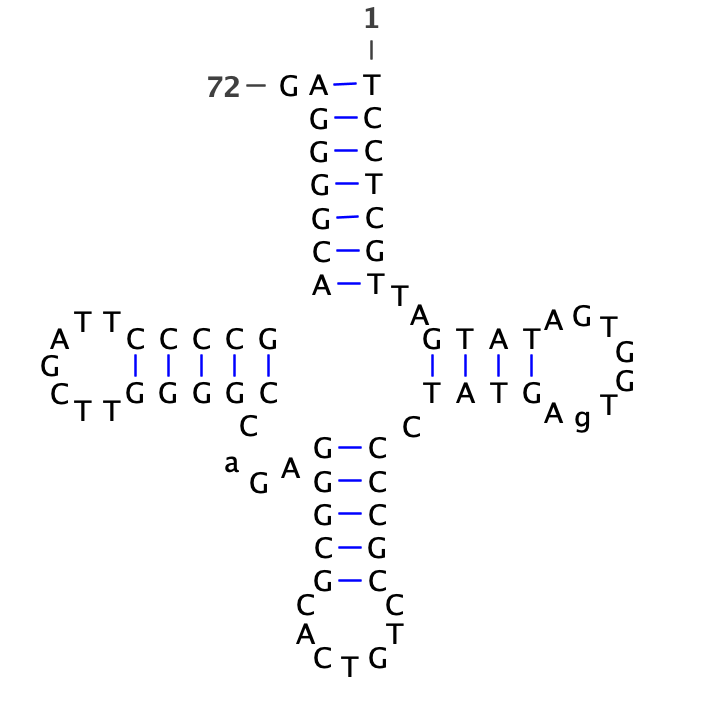
human Asp tRNA
human Leu tRNA
miRNA
miRNAs from miRBase are collected. The miRBase database is a searchable database of published miRNA sequences and annotation.
miRNA sequences and structures are located in /150T/zhangqf/GenomeAnnotation/miRNA
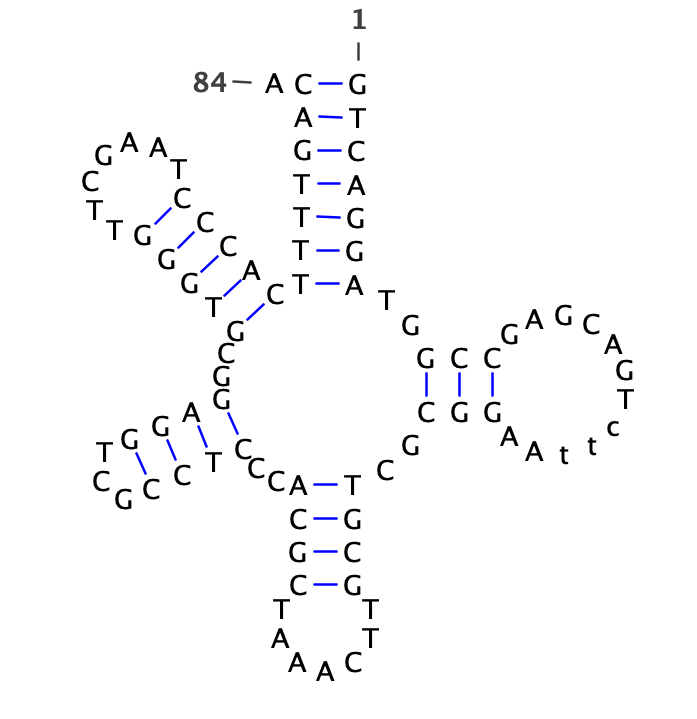
ccuuggccauguAAAAGUGCUUACAGUGCAGGUAGcuuuuugagaucuaCUGCAAUGUAAGCACUUCUUACauuaccaugg
((.(((..(((((((((((((((((.(((((.(((.((.....)).)))))))).))))))))))).))))))..))).))
snoRNA
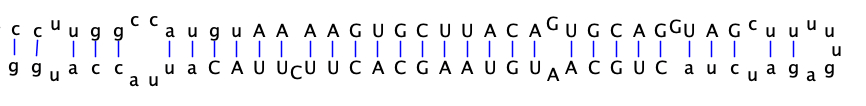
snoRNAs from snoatlas are collected. Small nucleolar RNAs (snoRNAs) are a class of non-coding RNAs that guide the post-transcriptional processing of other non-coding RNAs (mostly ribosomal RNAs), but have also been implicated in processes ranging from microRNA-dependent gene silencing to alternative splicing
snoRNA sequences are located in /150T/zhangqf/GenomeAnnotation/SnoRNA
GGGCTTGATAACTGTGCAAATGATTTTCTCAAATAGCATTGTGATGATGCCAAGAGCTCTGAATGCT
>HACA_67-1_P.troglodytes_(adopted_1)_135095584,135095720_-_hsa_HACA_67-5_97.81-100.74-189_AGAGTA_61_ACA_132
GCATGGTAATGGATTTATGGTGGGTCCTTCTCTCTGGGCCTCTCATAGTGTACCCATGCCAGAGTAAATGGCAGCCTCGAACCATTGCCCAGTCCCCTTACCTGTGGGCTGTGAGCACTGAAGGGGGTTGCACAGTG