Rfam
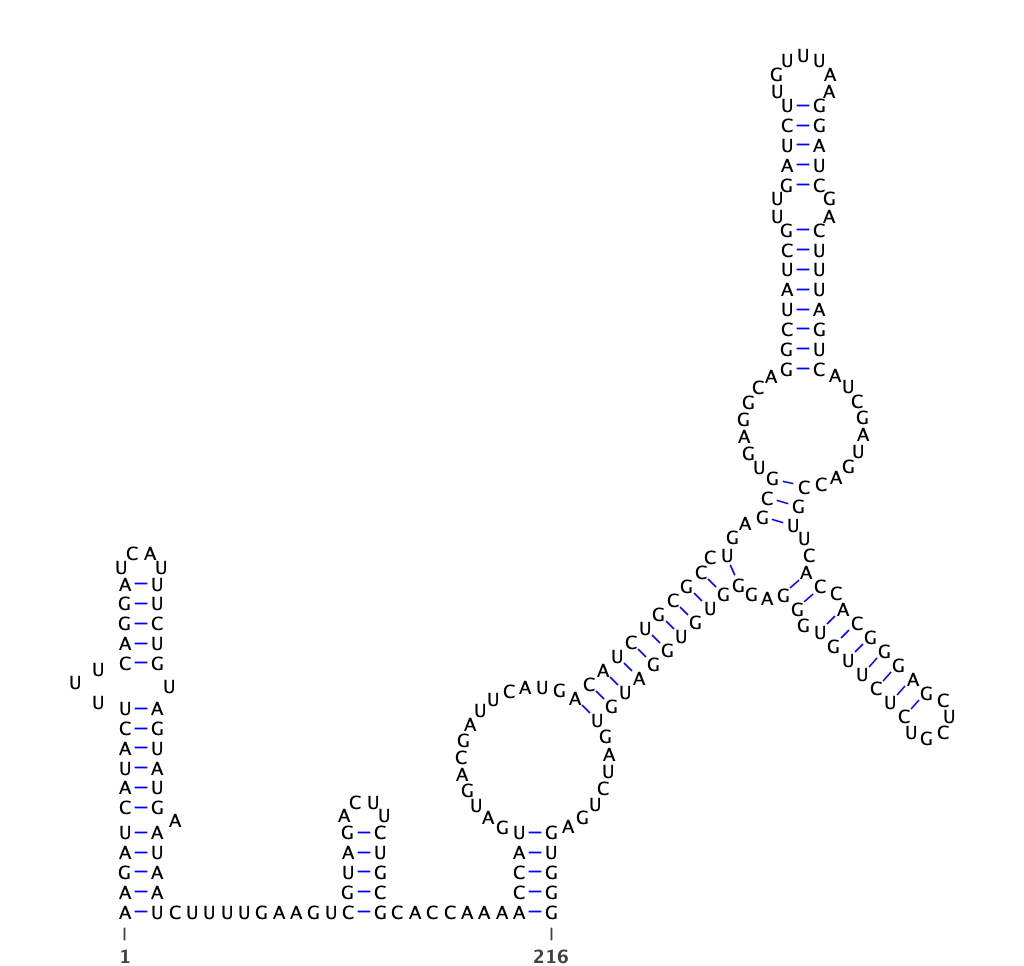
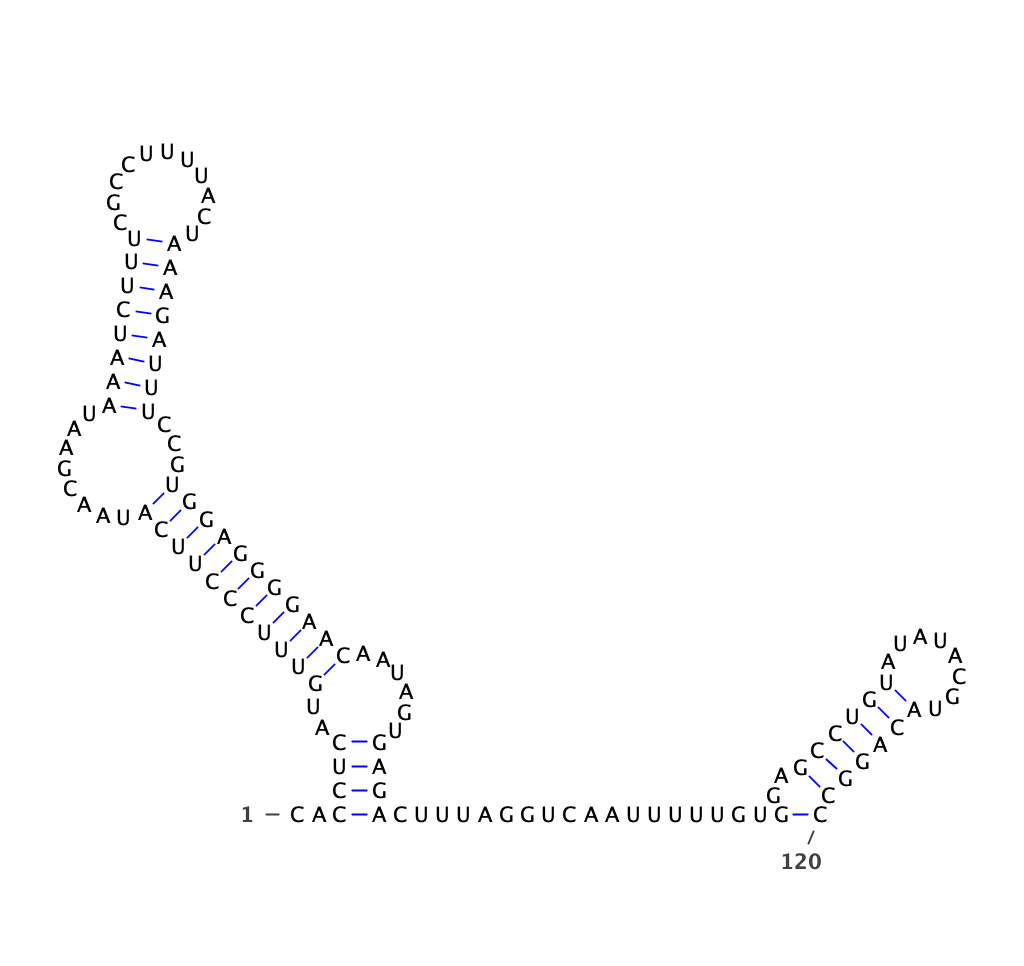
The Rfam database is a collection of RNA families, each represented by multiple sequence alignments, consensus secondary structures and covariance models (CMs). Conserved RNA structures (*.dot) of human have been parsed from multiple-alignment files (*.seed)
Rfam database is located in /150T/zhangqf/GenomeAnnotation/Rfam
seed
The seed alignments are hand curated and aligned using available sequence and structure data, and covariance models are built from these alignments using the INFERNAL v1.1.2 software suite (http://infernal.janelia.org). It is a .stockholm file
You can parse conserved structures for all kinds of species with this file
seed file is located in /150T/zhangqf/GenomeAnnotation/Rfam/14.1/Rfam.seed
human conserved structures
Conserved structures from human have been parsed with python script in Parsed_Structure
dot file is located in /150T/zhangqf/GenomeAnnotation/Rfam/Parsed_Structure/human.dot
U3
U5