
2024
2023
2022
2021
2020
2019
2018
2017
2016
Before2016
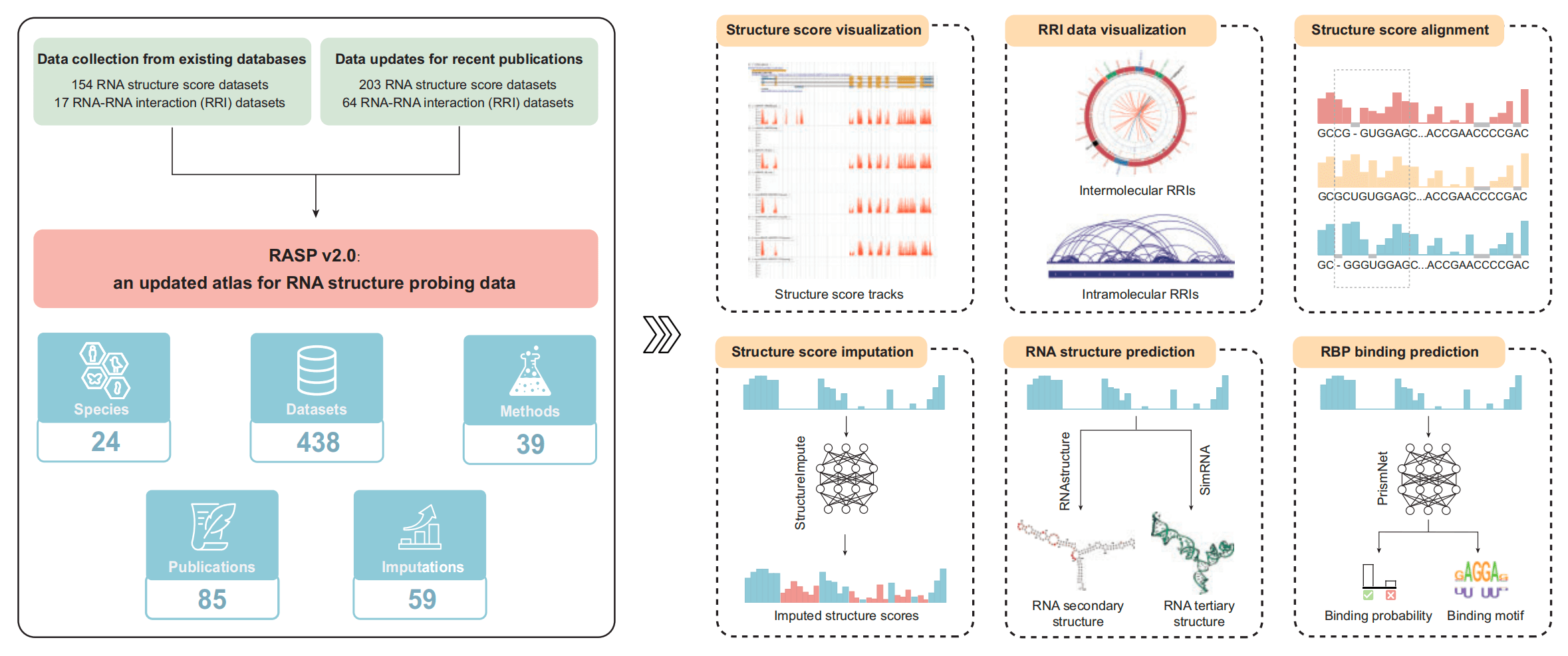
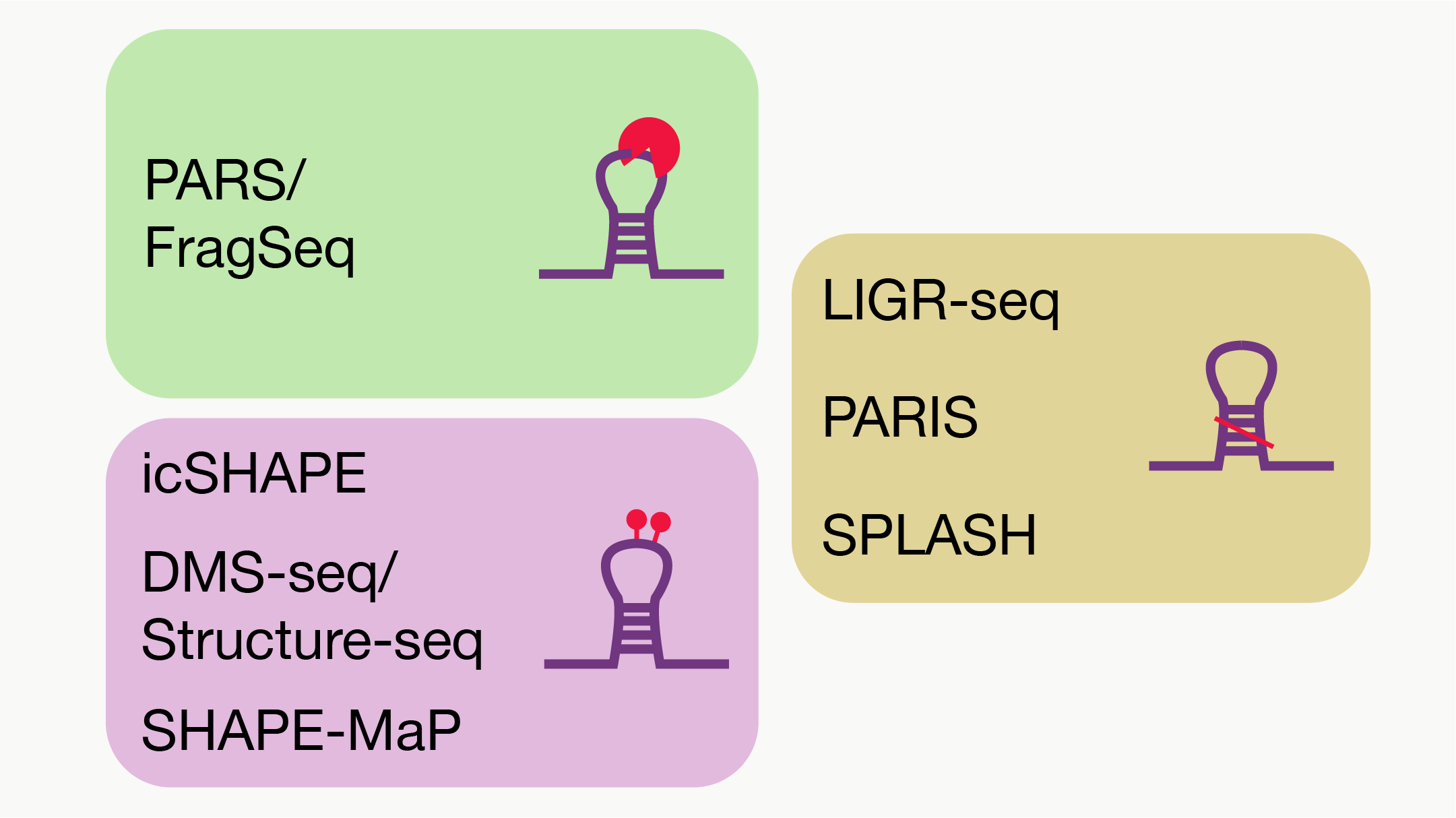
RASP v2.0:an updated atlas for RNA structure probing data
Kunting Mu*, Yuhan Fei*#, Yiran Xu, Qiangfeng Cliff Zhang#
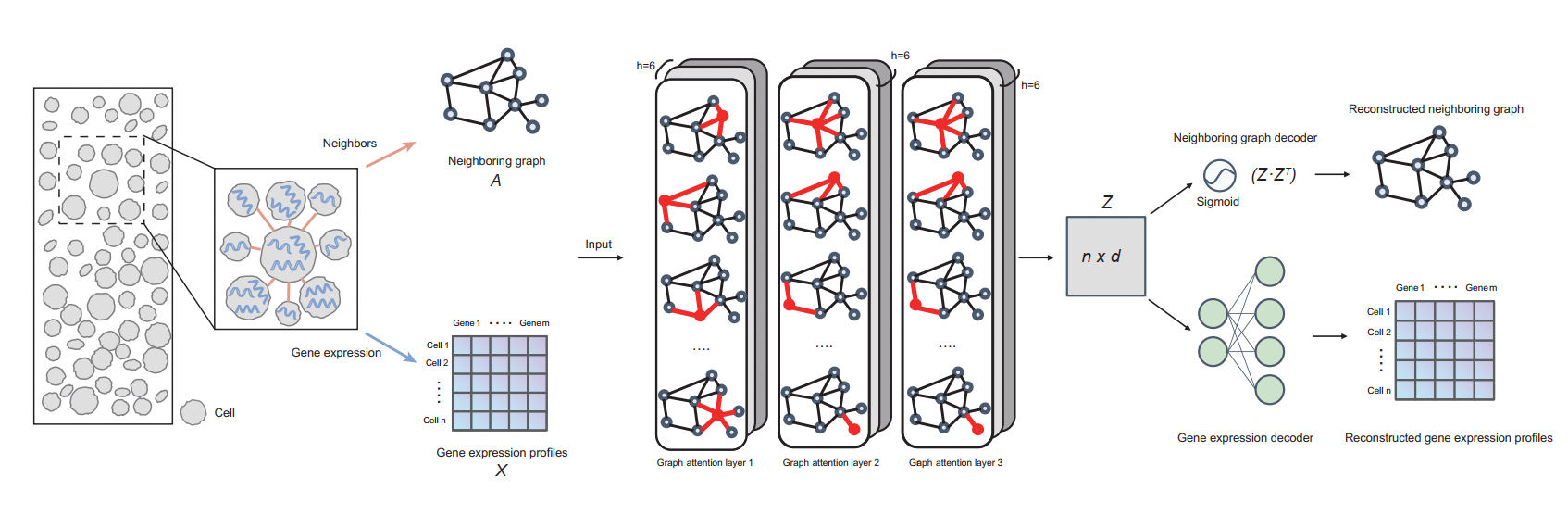
Tissue module discovery in single-cell-resolution spatial transcriptomics data via cell-cell interaction-aware cell embedding
Yuzhe Li, Jinsong Zhang, Xin Gao, Qiangfeng Cliff Zhang
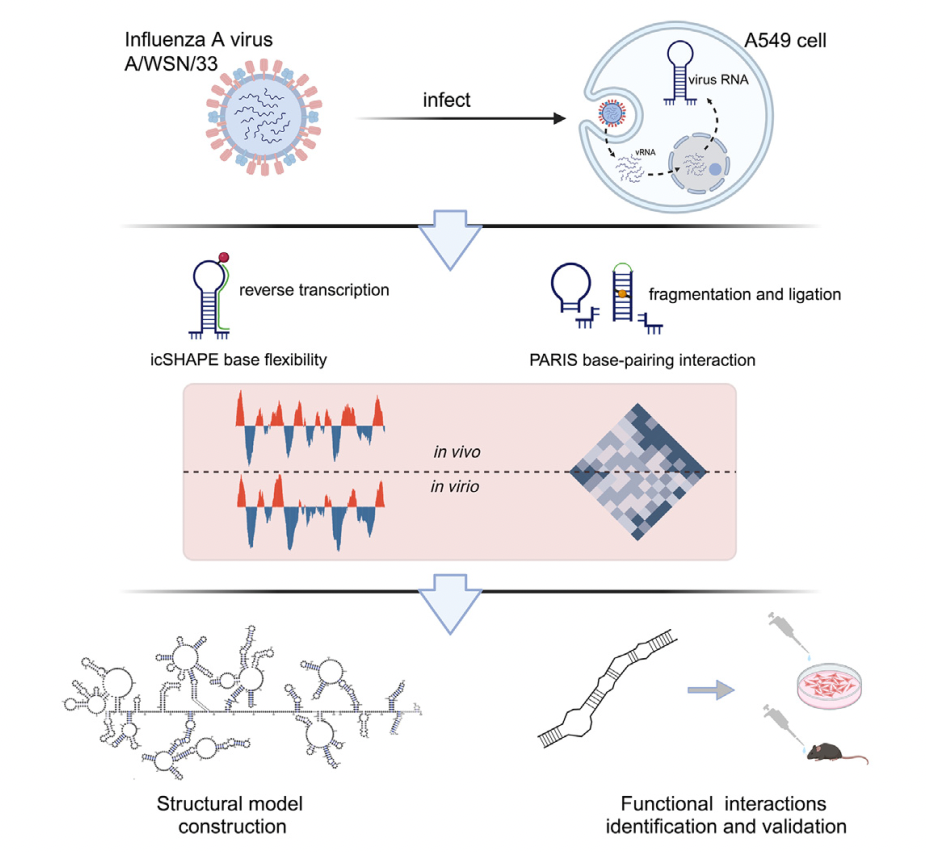
Mapping of the influenza A virus genome RNA structure and interactions reveals essential elements of viral replication
Rui Yang, Minglei Pan, Jiamei Guo, Yong Huang, Qiangfeng Cliff Zhang, Tao Deng, Jianwei Wang
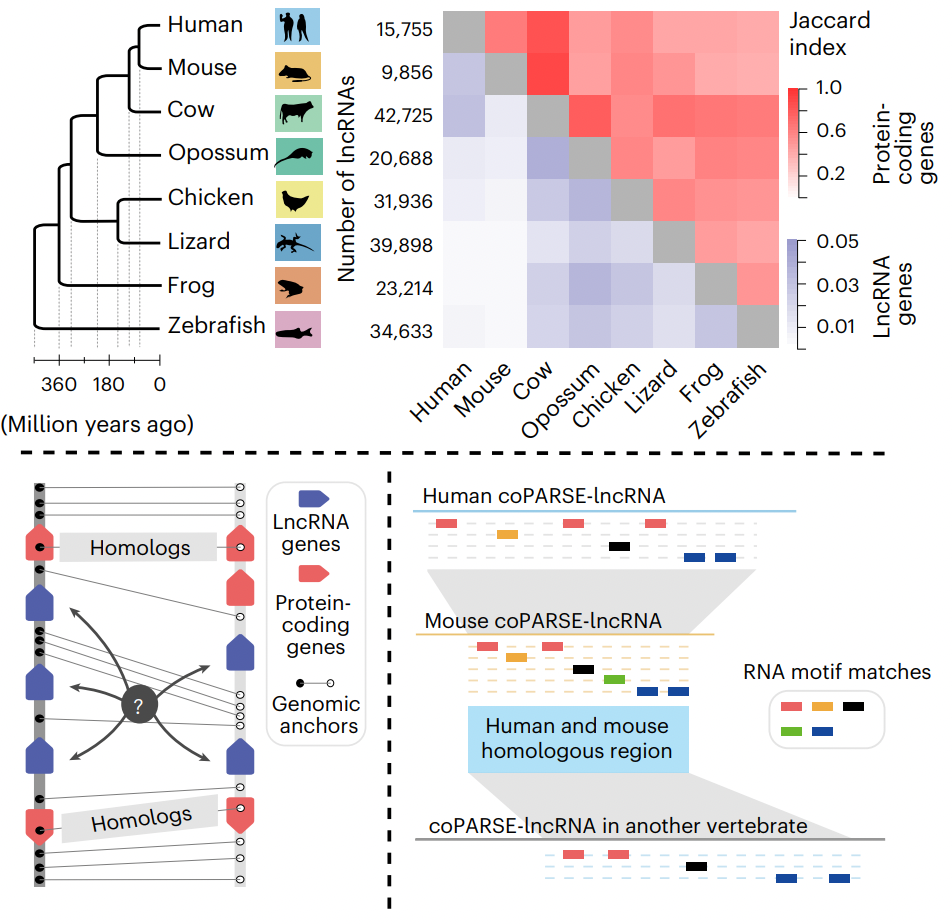
Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human
Wenze Huang, Tuanlin Xiong, Yuting Zhao, Jian Heng, Ge Han, Pengfei Wang, Zhihua Zhao, Ming Shi, Juan Li, Jiazhen Wang, Yixia Wu, Feng Liu, Jianzhong Jeff Xi, Yangming Wang & Qiangfeng Cliff Zhang
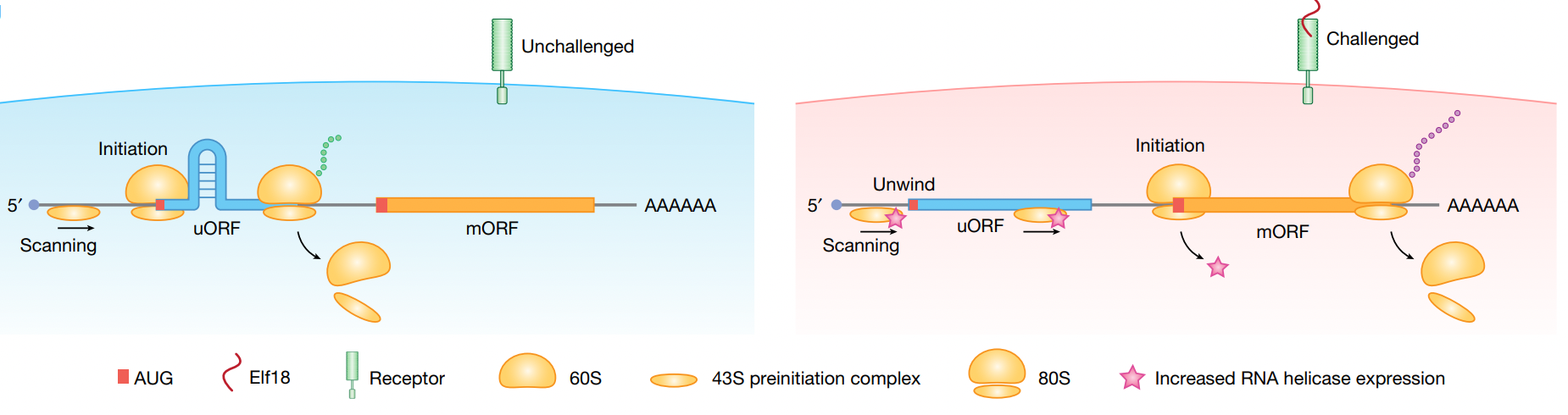
Pervasive downstream RNA hairpins dynamically dictate start-codon selection
Yezi Xiang, Wenze Huang, Lianmei Tan, Tianyuan Chen, Yang He, Patrick S. Irving, Kevin M. Weeks, Qiangfeng Cliff Zhang & Xinnian Dong

RNA structure determination: From 2D to 3D
Jie Deng, Xianyang Fang#, Lin Huang#, Shanshan Li, Lilei Xu, Keqiong Ye#, Jinsong Zhang, Kaiming Zhang# and Qiangfeng Cliff Zhang#
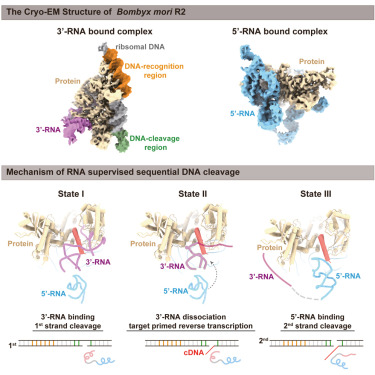
Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon
Pujuan Deng*, Shun-Qing Tan*, Qi-Yu Yang, Han-Zhou Zhu, Lei Sun, Zhangbin Bao, Yi Lin, Qiangfeng Cliff Zhang, Jia Wang# and Jun-Jie Gogo Liu#
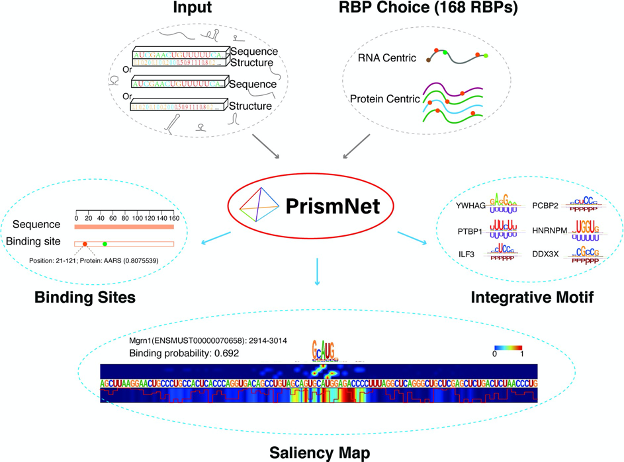
PrismNet: predicting protein–RNA interaction using in vivo RNA structural information
Yiran Xu*, Jianghui Zhu*, Wenze Huang*, Kui Xu, Rui Yang, Qiangfeng Cliff Zhang# and Lei Sun#
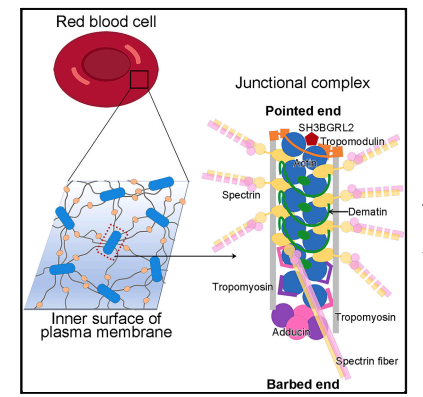
Structural basis of membrane skeleton organization in red blood cells
Ningning Li, Siyi Chen, Kui Xu, Meng-Ting He, Meng-Qiu Dong, Qiangfeng Cliff Zhang, Ning Gao
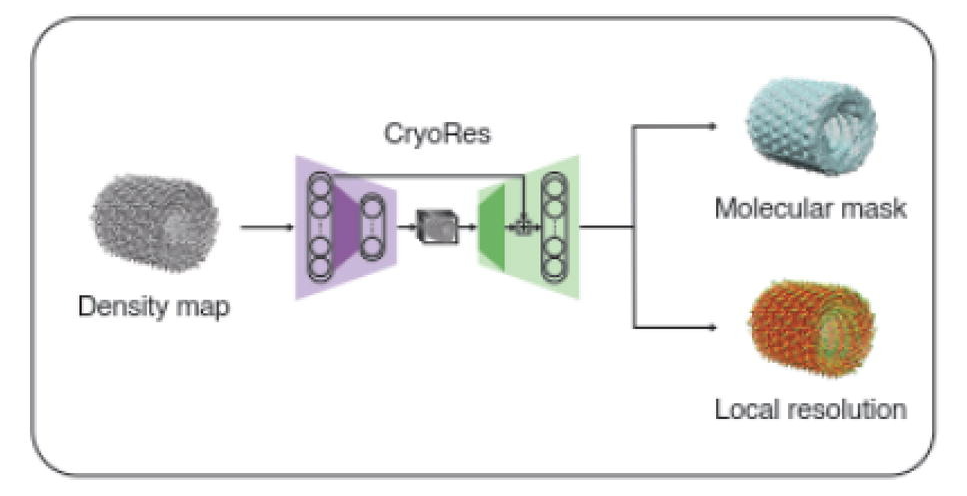
CryoRes: Local Resolution Estimation of Cryo-EM Density Maps by Deep Learning
Muzhi Dai, Zhuoer Dong, Kui Xu, Qiangfeng Cliff Zhang
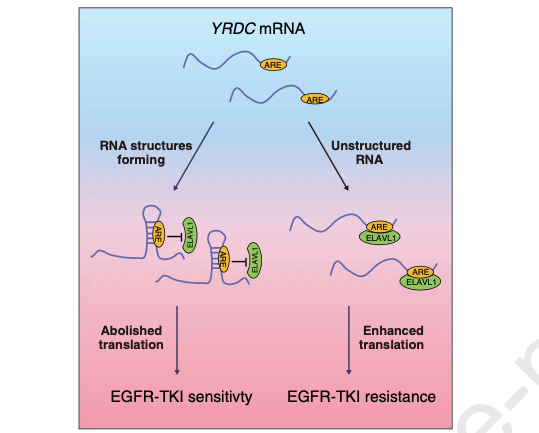
RNA Structural Dynamics Modulate EGFR-TKI Resistance Through Controlling YRDC Translation in NSCLC Cells
Boyang Shi, Ke An, Yueqin Wang, Yuhan Fei, Caixia Guo, Qiangfeng Cliff Zhang, Yun-Gui Yang, Xin Tian, Quancheng Kan
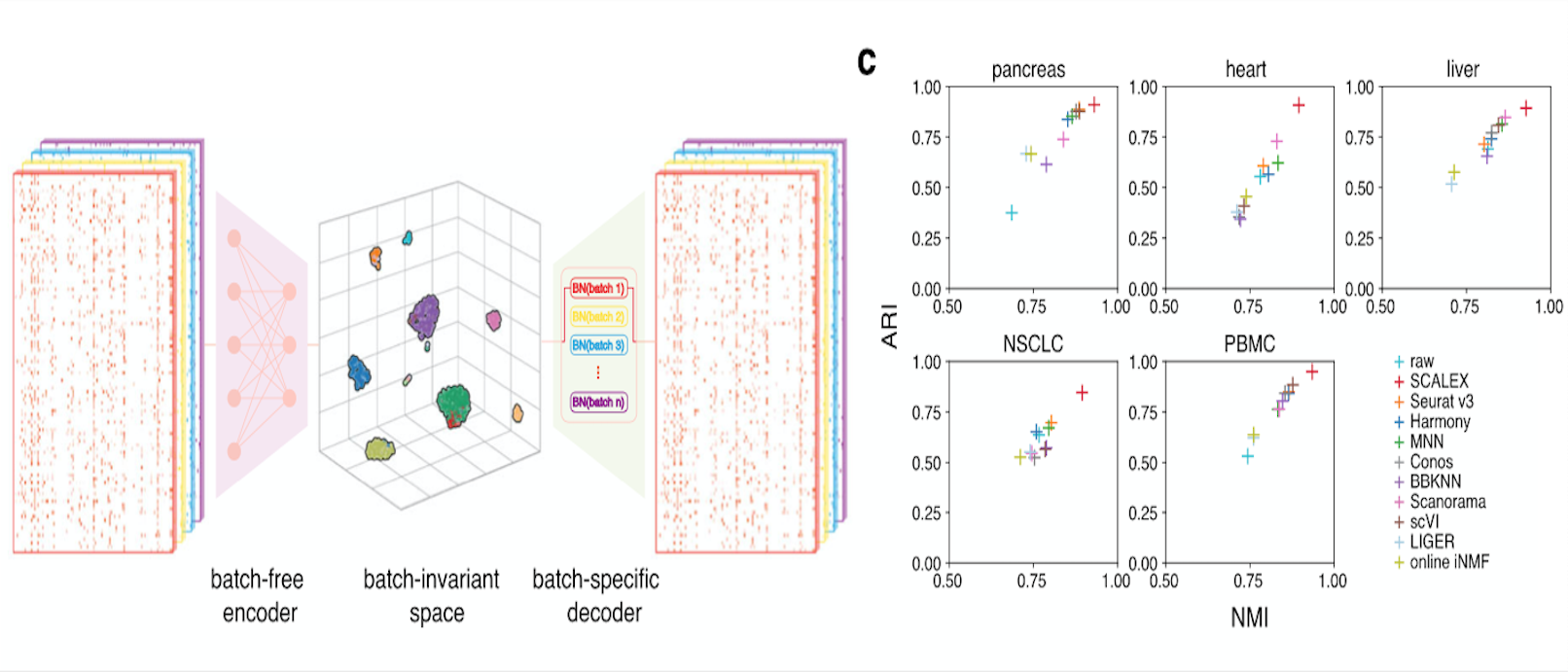
Online single-cell data integration through projecting heterogeneous datasets into a common cell-embedding space
Lei Xiong, Kang Tian, Yuzhe Li, Weixi Ning, Xin Gao, Qiangfeng Cliff Zhang
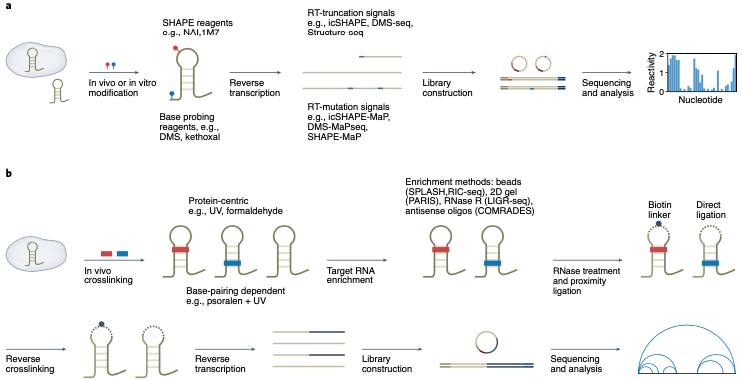
Advances and opportunities in RNA structure experimental determination and computational modeling
Jinsong Zhang, Yuhan Fei, Lei Sun, Qiangfeng Cliff Zhang
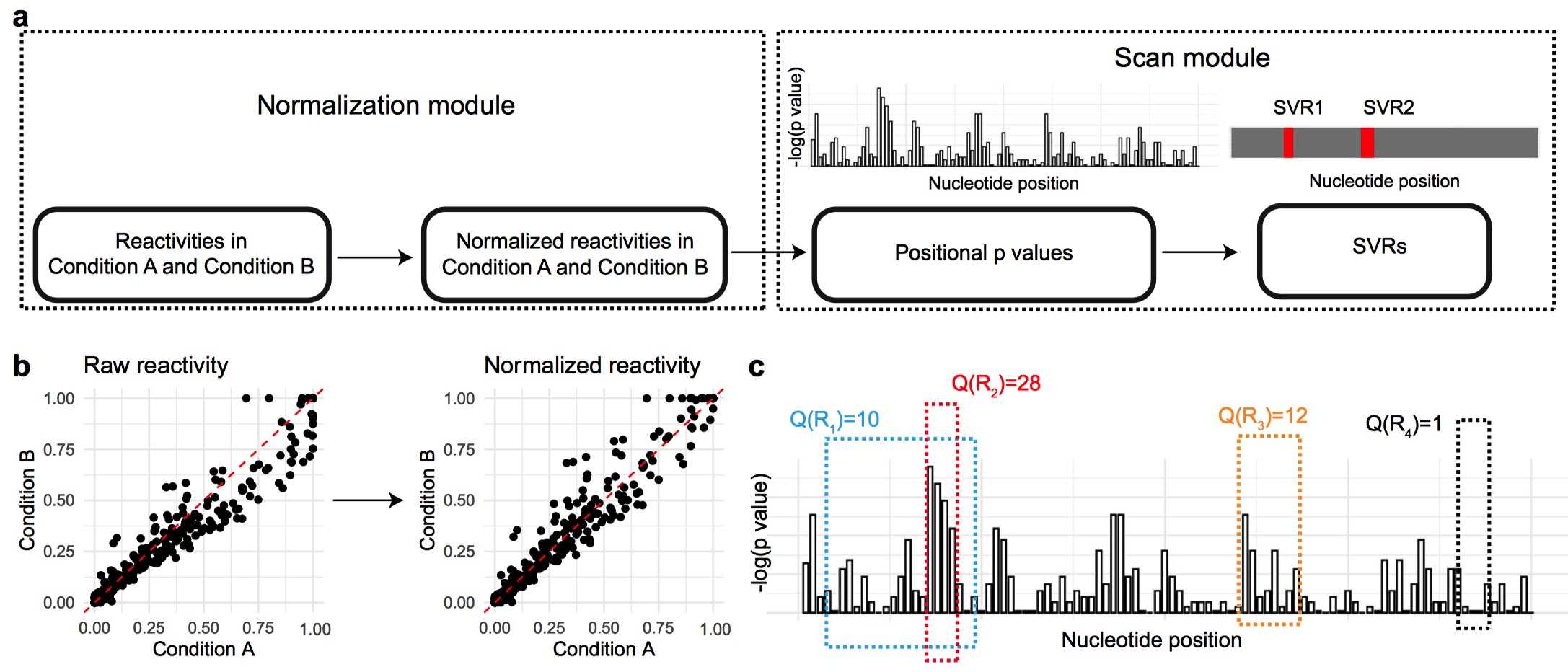
Differential analysis of RNA structure probing experiments at nucleotide resolution: uncovering regulatory functions of RNA structure
Bo Yu, Pan Li, Qiangfeng Cliff Zhang, Lin Hou
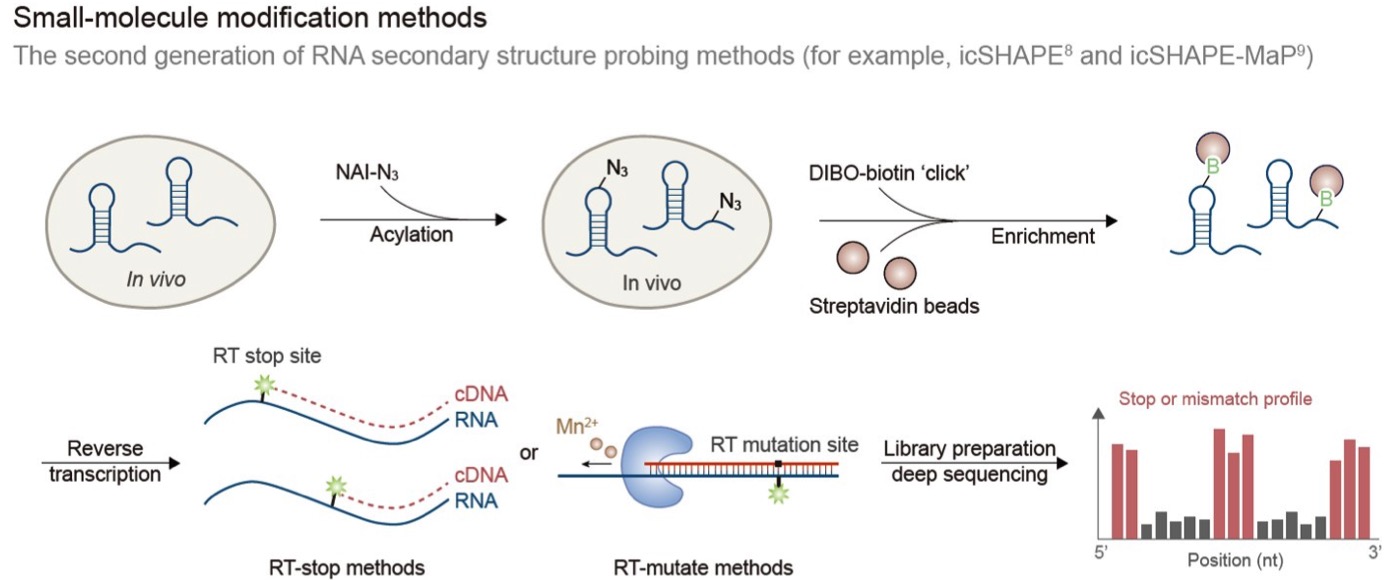
Recent advances in RNA structurome
Bingbing Xu, Yanda Zhu, Changchang Cao, Hao Chen, Qiongli Jin, Guangnan Li, Junfeng Ma, Siwy Ling Yang, Jieyu Zhao, Jianghui Zhu, Yiliang Ding, Xianyang Fang, Yongfeng Jin, Chun Kit Kwok, Aiming Ren, Yue Wan, Zhiye Wang, Yuanchao Xue, Huakun Zhang, Qiangfeng Cliff Zhang, Yu Zhou
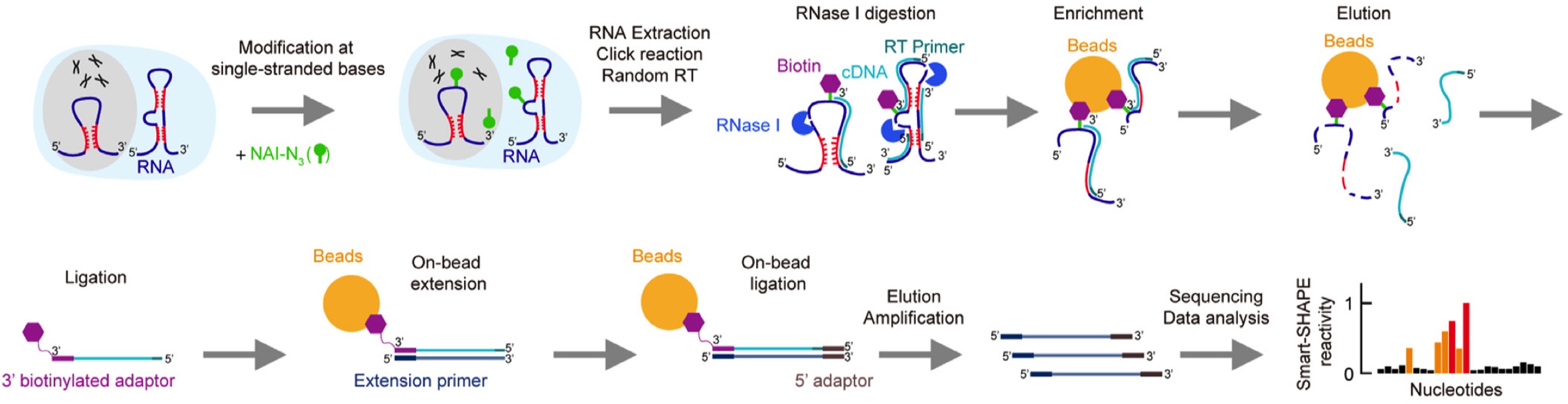
An ultra low-input method for global RNA structure probing uncovers Regnase-1-mediated regulation in macrophages
Meiling Piao, Pan Li, Xiaomin Zeng, Xiwen Wang, Lan Kang, jinsong Zhang, Yifan Wei, Shaojun Zhang, Lei Tang, Jianghui Zhu, Chun Kit Kwok, Xiaoyu Hu, Qiangfeng Cliff Zhang
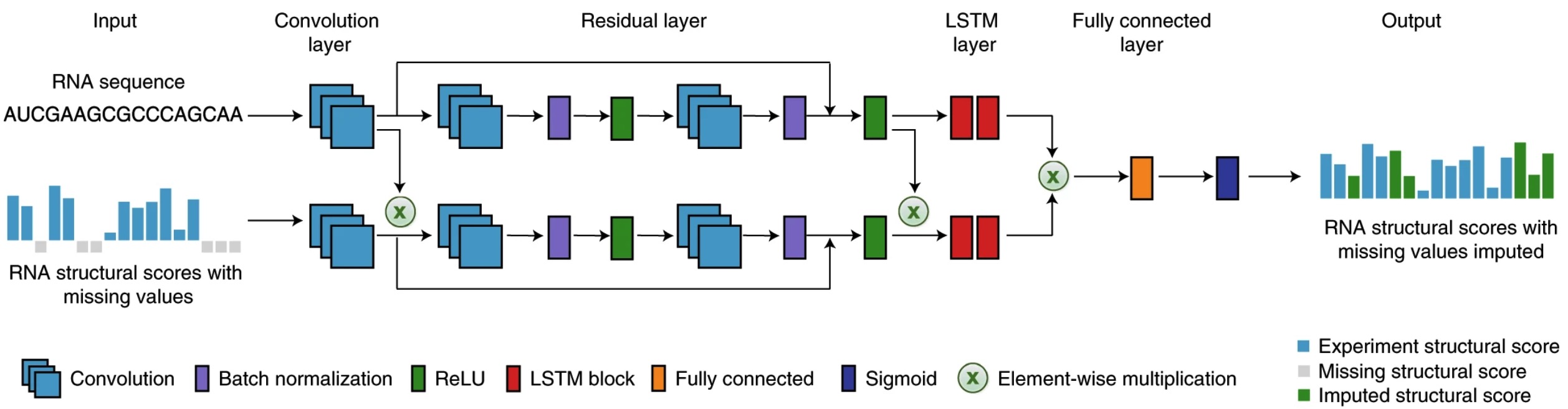
A deep learning method for recovering missing signals in transcriptome-wide RNA structure profiles from probing experiments
Jing Gong*, Kui Xu*, Ziyuan Ma, Zhi John Lu and Qiangfeng Cliff Zhang#
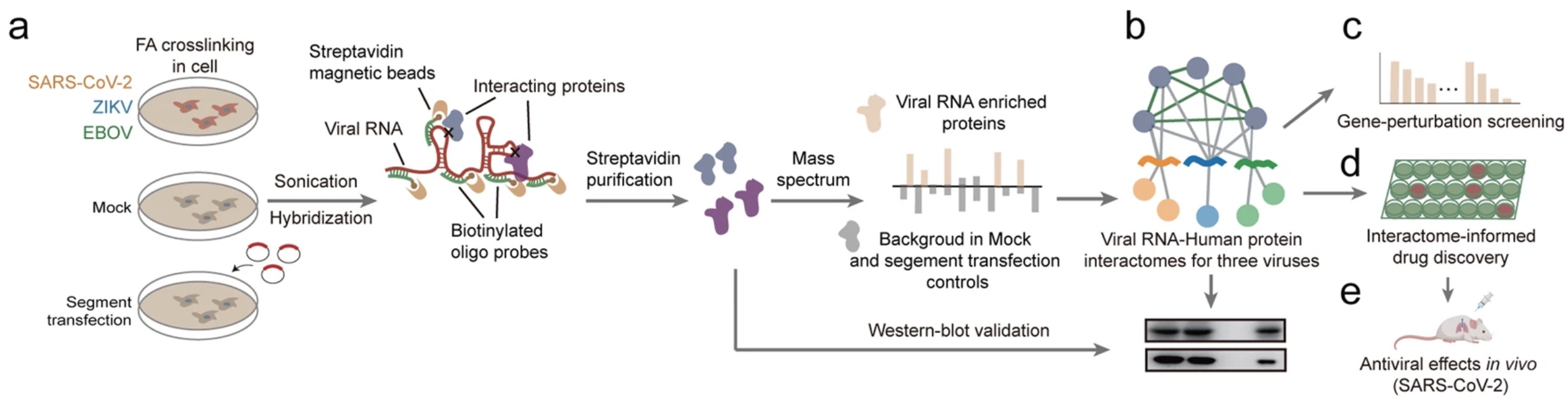
Comparison of viral RNA–host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2
Shaojun Zhang*, Wenze Huang*, Lili Ren*, Xiaohui Ju*, Mingli Gong*, Jian Rao*, Lei Sun, Pan Li, Qiang Ding#, Jianwei Wang# and Qiangfeng Cliff Zhang#
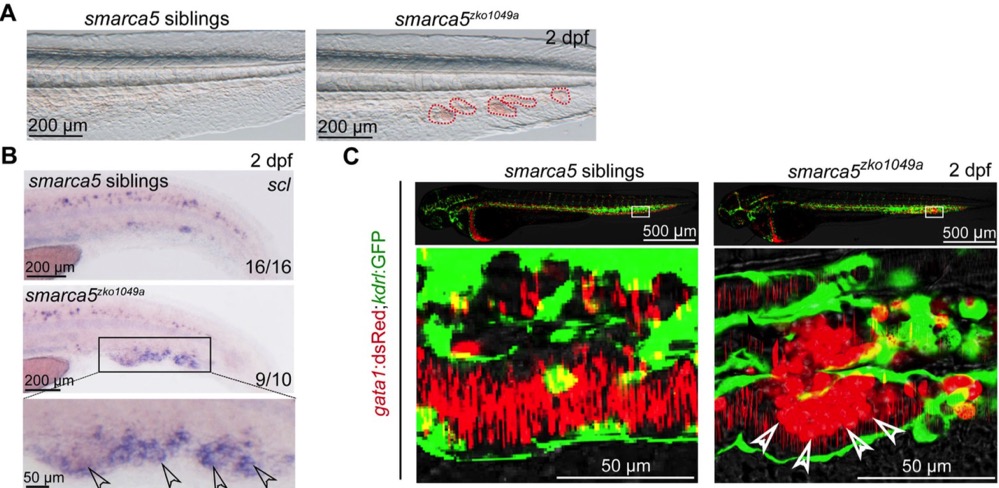
The chromatin-remodeling enzyme Smarca5 regulates erythrocyte aggregation via Keap1-Nrf2 signaling
Yanyan Ding*, Yuzhe Li*, Ziqian Zhao*, Qiangfeng Cliff Zhang and Feng Liu#
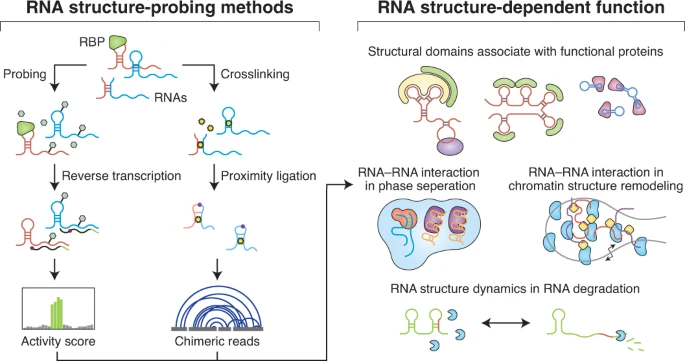
RNA structure probing uncovers RNA structure-dependent biological functions
Xiwen Wang*, Chuxiao Liu*, Lingling Chen# and Qiangfeng Cliff Zhang#
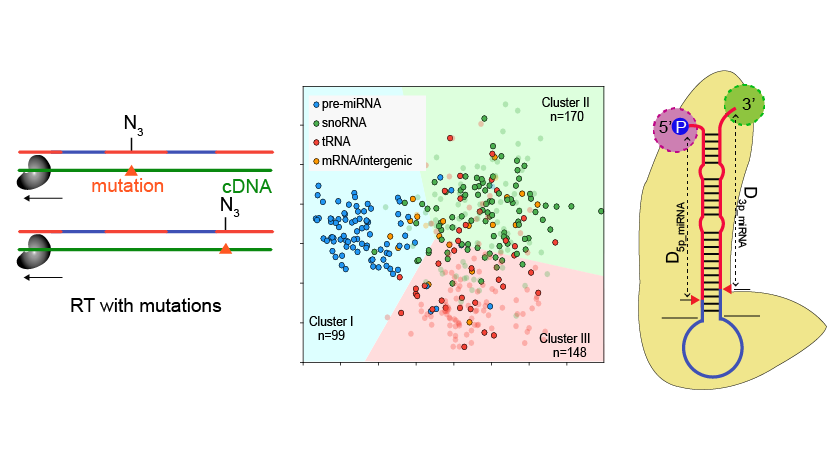
RNA structure probing reveals the structural basis of Dicer binding and cleavage
Qingjun Luo*, Jinsong Zhang*, Pan Li*, Qing Wang, Yue Zhang, Biswajoy Roy-Chaudhuri, Jianpeng Xu, Mark A. Kay#, Qiangfeng Cliff Zhang#
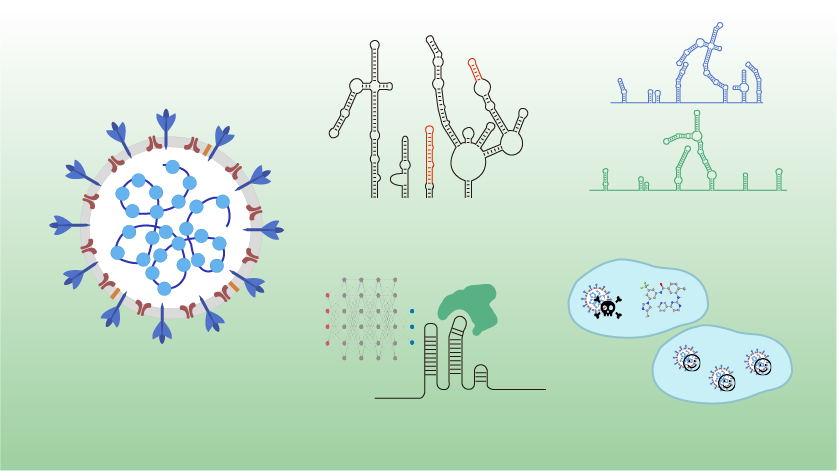
In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs
Lei Sun*,Pan Li*,Xiaohui Ju*,Jian Rao*,Wenze Huang*,Lili Ren,Shaojun Zhang,Tuanlin Xiong,Kui Xu,Xiaolin Zhou,Mingli Gong,Eric Miska,Qiang Ding#,Jianwei Wang#,Qiangfeng Cliff Zhang#
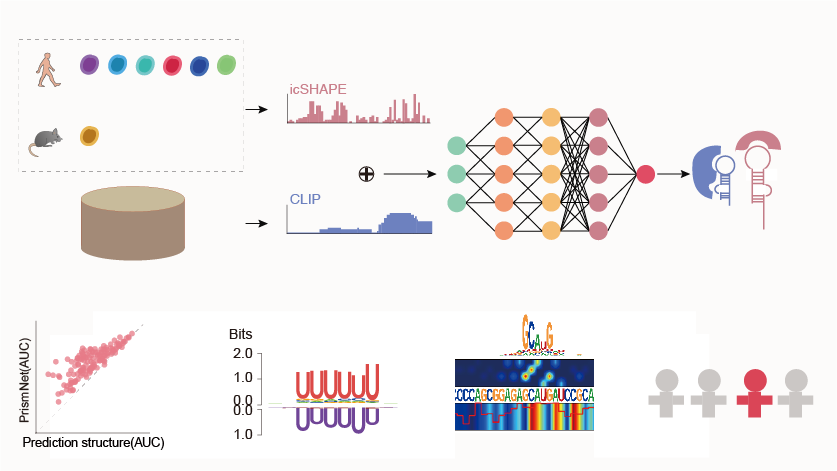
Predicting dynamic cellular protein–RNA interactions by deep learning using in vivo RNA structures
Lei Sun*, Kui Xu*, Wenze Huang*, Yucheng T. Yang*, Pan Li, Lei Tang, Tuanlin Xiong, Qiangfeng Cliff Zhang#

Structure of the activated human minor spliceosome
Rui Bai*, Ruixue Wan*#, Lin Wang, Kui Xu, Qiangfeng Cliff Zhang, Jianlin Lei, Yigong Shi#

IRIS: A method for predicting in vivo RNA secondary structures using PARIS data
Jianyu Zhou, Pan Li, Wanwen Zeng, Wenxiu Ma, Zhipeng Lu, Rui Jiang, Qiangfeng Cliff Zhang#, Tao Jiang#
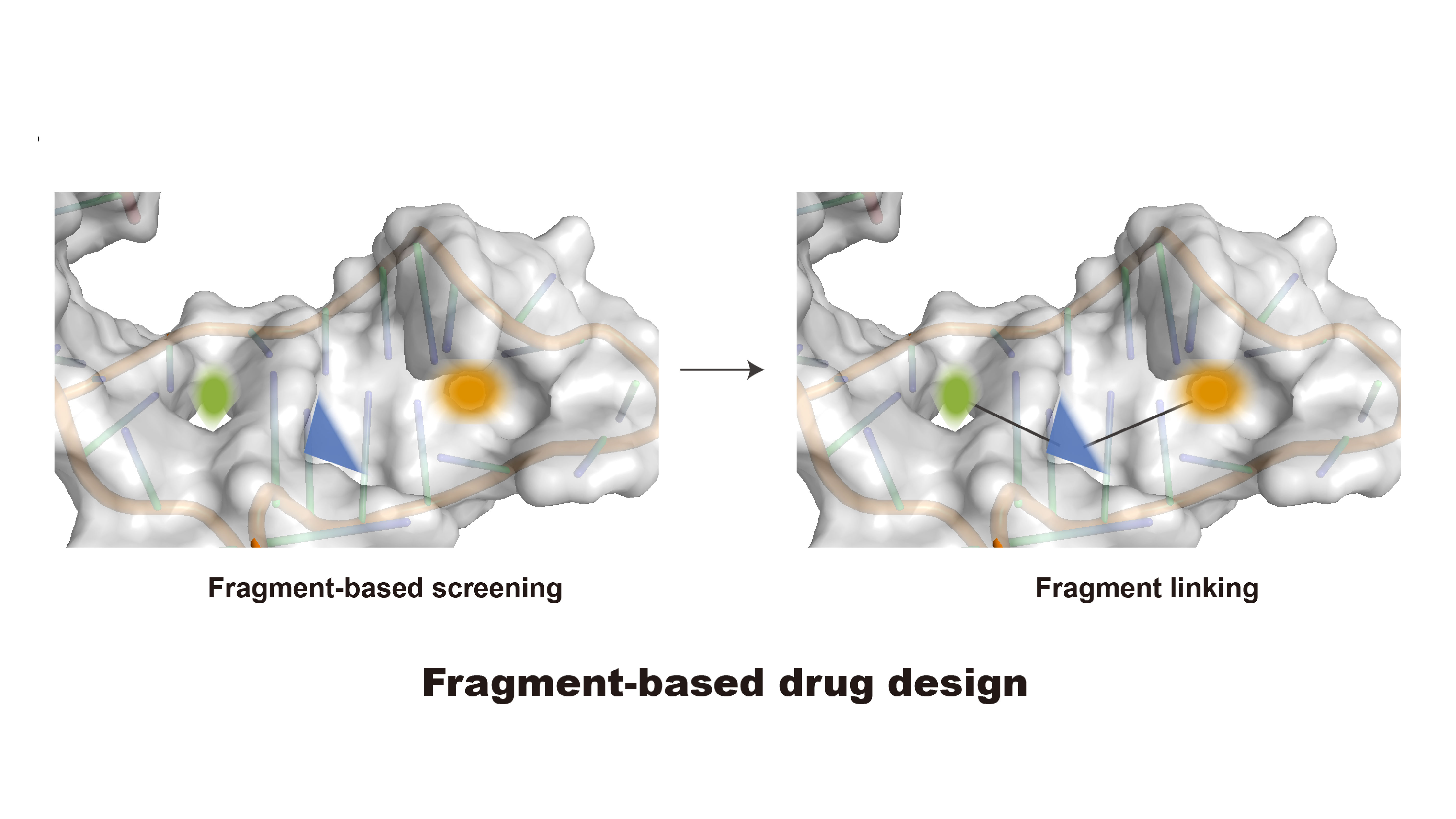
Targeting RNA structures in diseases with small molecules
Yanqiu Shao and Qiangfeng Cliff Zhang
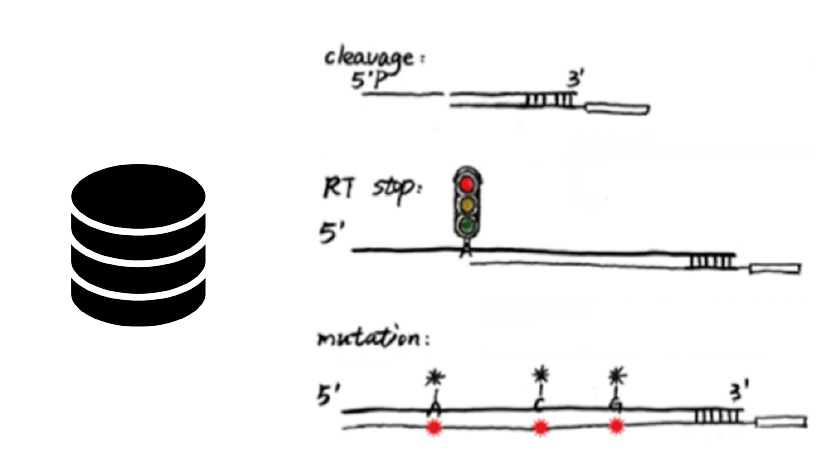
RASP: an atlas of transcriptome-wide RNA secondary structure probing data
Pan Li*, Xiaolin Zhou*, Kui Xu and Qiangfeng Cliff Zhang#
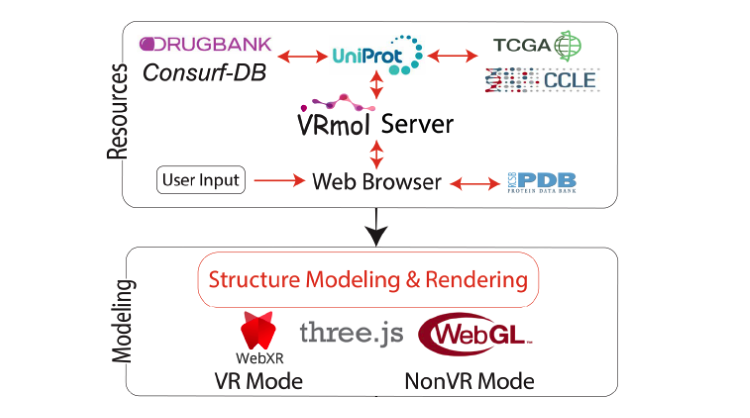
VRmol: an Integrative Web-Based Virtual Reality System to Explore Macromolecular Structure
Kui Xu*, Nan Liu*, Jingle Xu, Chunlong Guo, Lingyun Zhao, Hong-Wei Wang, Qiangfeng Cliff Zhang#
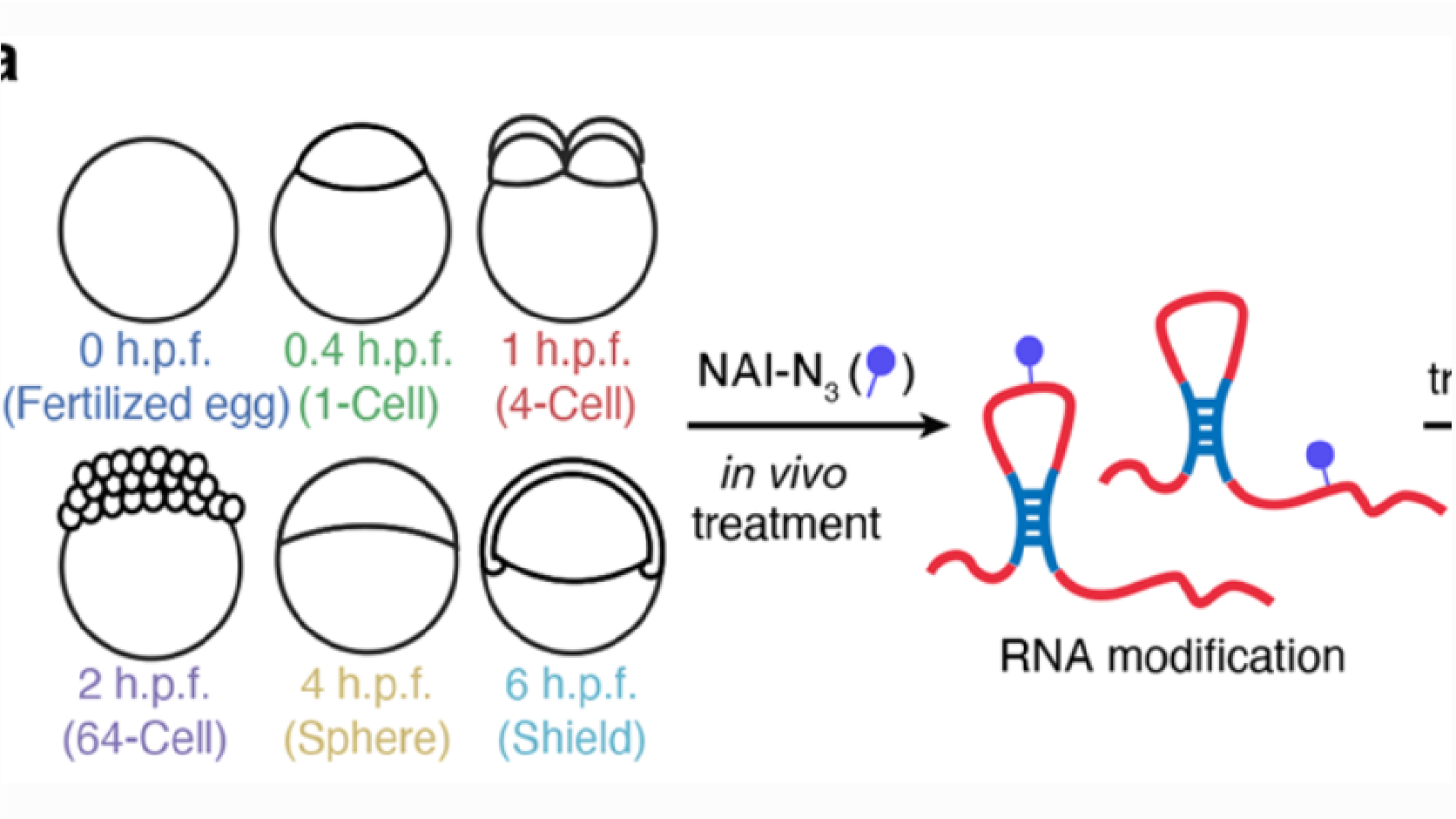
RNA structural dynamics regulate early embryogenesis through controlling transcriptome fate and function
Boyang Shi*, Jinsong Zhang*, Jian Heng*, Jing Gong*, Ting Zhang, Pan Li, Bao-Fa Sun, Ying Yang, Ning Zhang, Yong-Liang Zhao, Hai-Lin Wang, Feng Liu#, Qiangfeng Cliff Zhang# & Yun-Gui Yang#

U1 snRNP regulates chromatin retention of noncoding RNAs
Yafei Yin, J. Yuyang Lu, Xuechun Zhang, Wen Shao, Yanhui Xu, Pan Li, Yantao Hong,Li Cui, Ge Shan, Bin Tian, Qiangfeng Cliff Zhang & Xiaohua Shen#
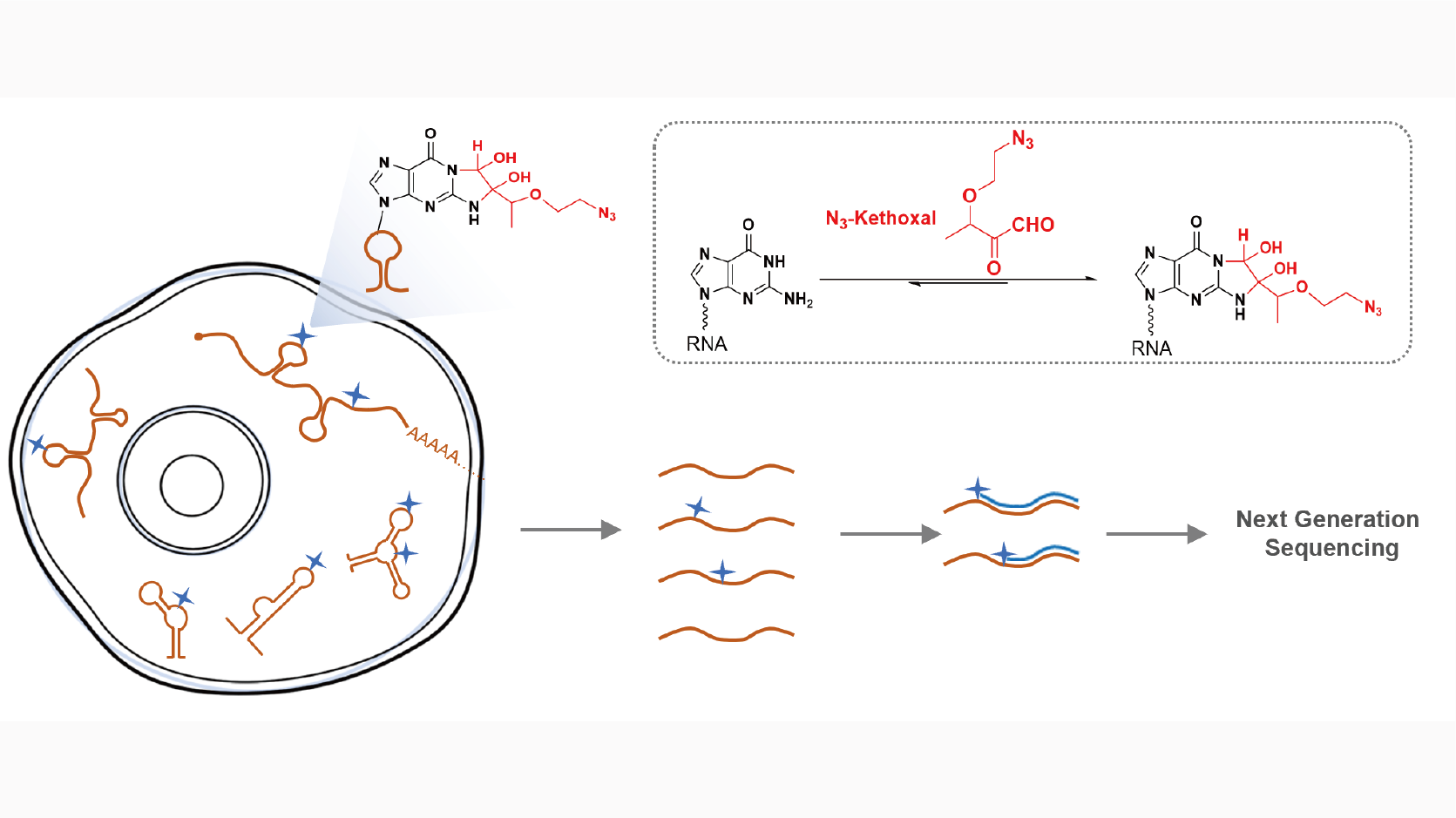
Keth-seq for transcriptome-wide RNA structure mapping
Xiaocheng Weng*, Jing Gong*, Yi Chen*, Tong Wu*, Fang Wang, Shixi Yang , Yushu Yuan, Guanzheng Luo, Kai Chen, Lulu Hu, Honghui Ma, Pingluan Wang, Qiangfeng Cliff Zhang#, Xiang Zhou#, Chuan He#

A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice
Hak Kyun Kim, Jianpeng Xu, Kirk Chu, Hyesuk Park, Hagoon Jang, Pan Li, Paul N. Valdmanis, Qiangfeng Cliff Zhang, Mark A. Kay

icSHAPE-pipe: A comprehensive toolkit for icSHAPE data analysis and evaluation
Pan Li, Ruoyao Shi, Qiangfeng Cliff Zhang
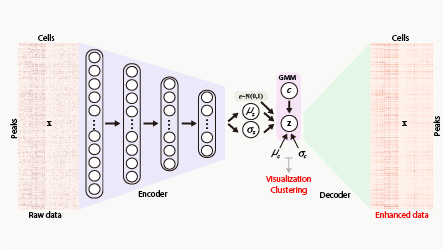
SCALE method for single-cell ATAC-seq analysis via latent feature extraction
Lei Xiong, Kui Xu, Kang Tian, Yanqiu Shao, Lei Tang, Ge Gao, Michael Zhang, Tao Jiang, Qiangfeng Cliff Zhang

Multivalent m6A motifs promote phase separation of YTHDF proteins
Yifei Gao, Gaofeng Pei, Dongxue Li, Ru Li, Yanqiu Shao, Qiangfeng Cliff Zhang and Pilong Li

Mapping In Vivo RNA Structures and Interactions
Jieyu Zhao*, Xingyang Qian*, Pui Yan Yeung*, Qiangfeng Cliff Zhang#, and Chun Kit Kwok#
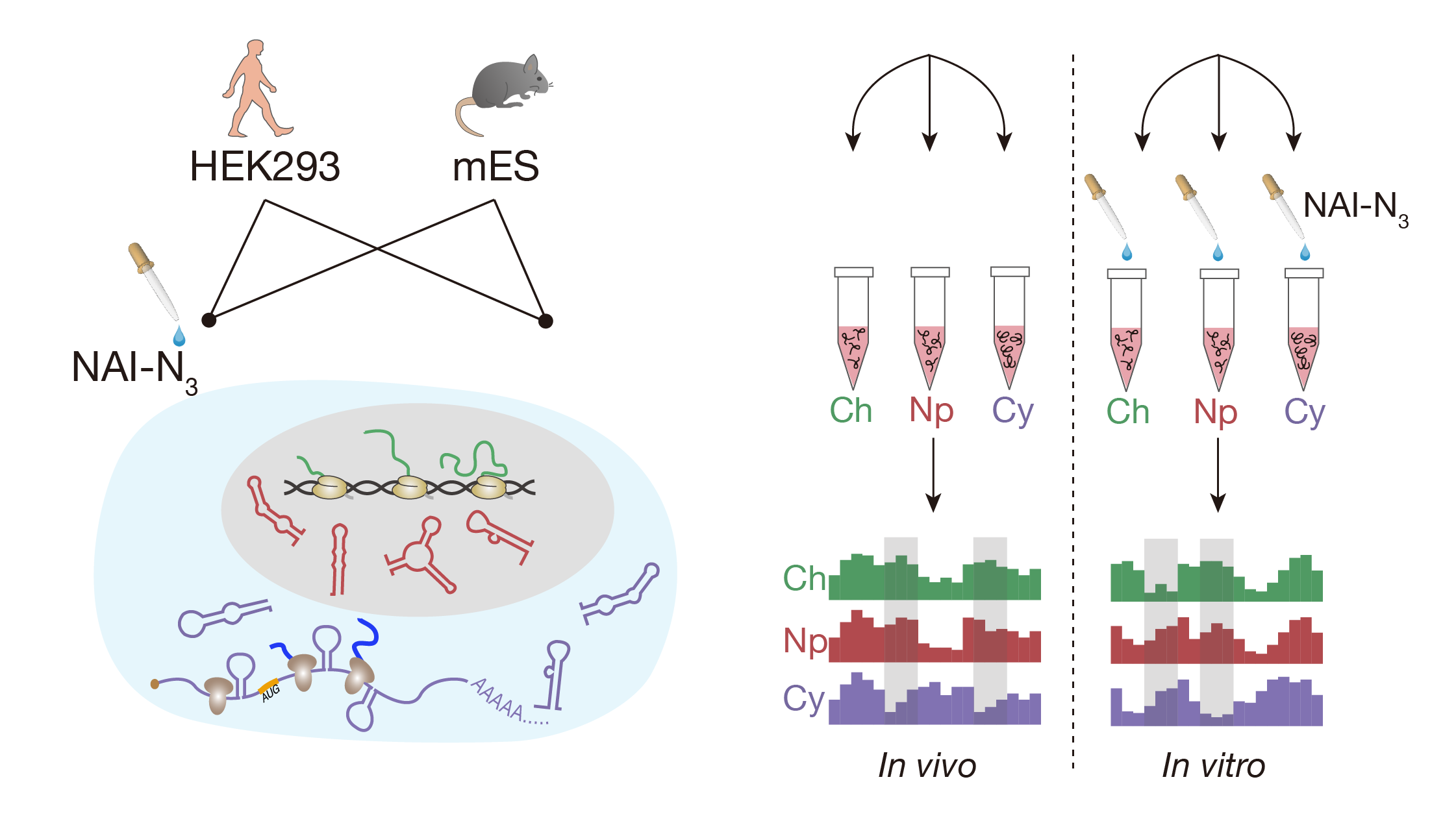
RNA structure maps across mammalian cellular compartments
Lei Sun*, Furqan M. Fazal*, Pan Li*, James P. Broughton, Byron Lee, Lei Tang, Wenze Huang, Eric T. Kool, Howard Y. Chang# and Qiangfeng Cliff Zhang#
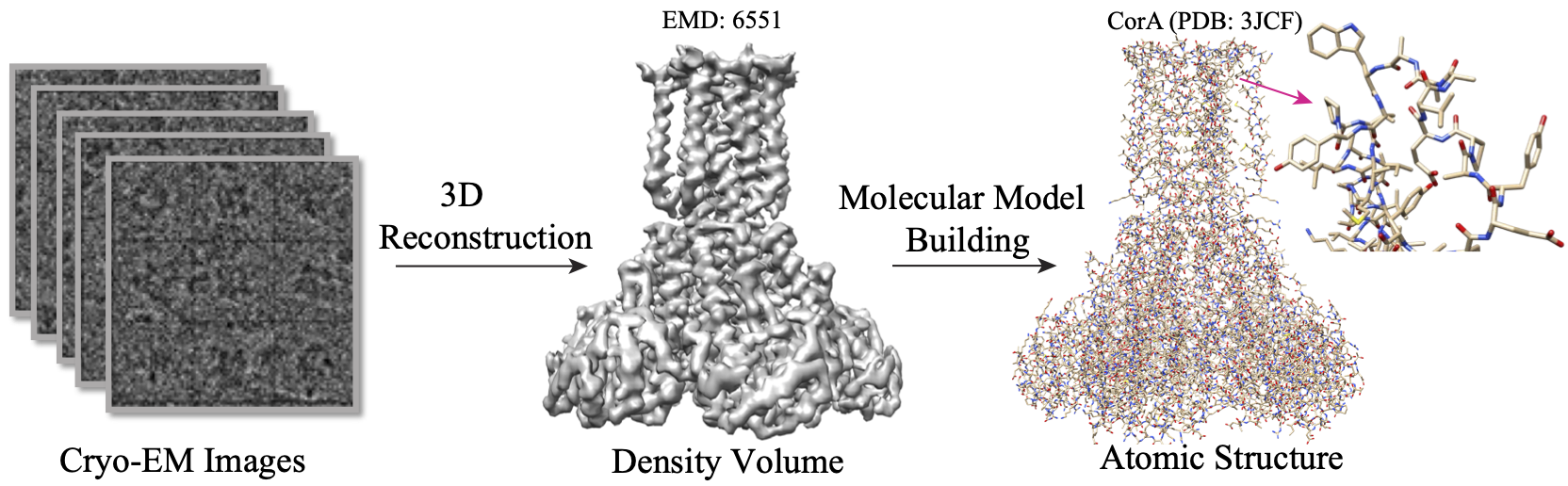
A²-Net: Molecular Structure Estimation from Cryo-EM Density Volumes
Kui Xu, Zhe Wang, Jianping Shi, Hongsheng Li, Qiangfeng Cliff Zhang
Proceedings of the AAAI Conference on Artificial Intelligence 33 (01), 1230-1237(Project)(arXiv)(PDF)

Investigation of RNA-RNA Interactions Using the RISE
Yanyan Ju*, Jing Gong*, Yucheng T. Yang#, and Qiangfeng Cliff Zhang#
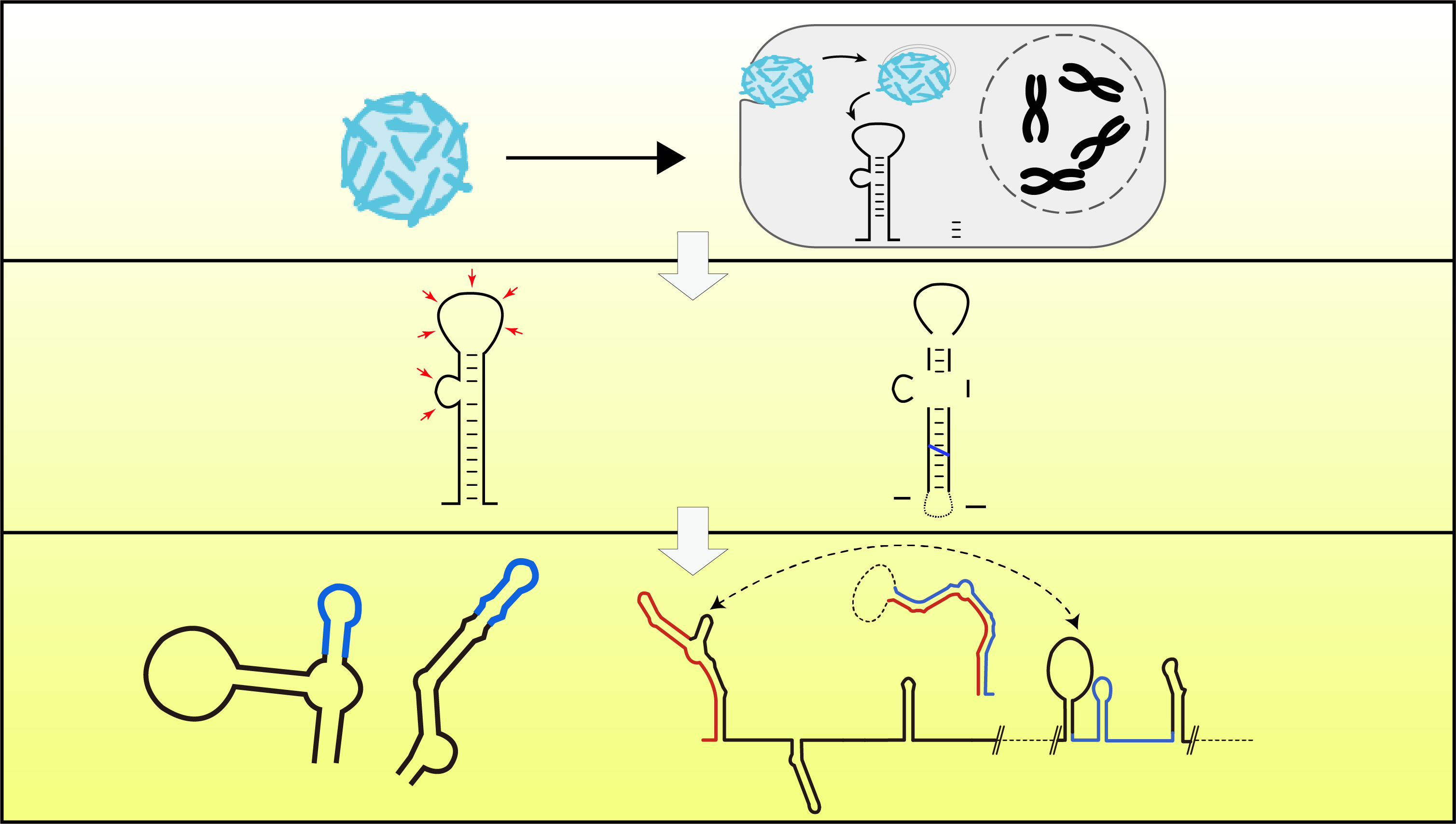
Integrative Analysis of Zika Virus Genome RNA Structure Reveals Critical Determinants of Viral Infectivity
Pan Li*, Yifan Wei*, Miao Mei*, Lei Tang*, Lei Sun, Wenze Huang, Jianyu Zhou, Chunlin Zou, Shaojun Zhang, Cheng-Feng Qin, Tao Jiang, Jianfeng Dai, Xu Tan#, and Qiangfeng Cliff Zhang#
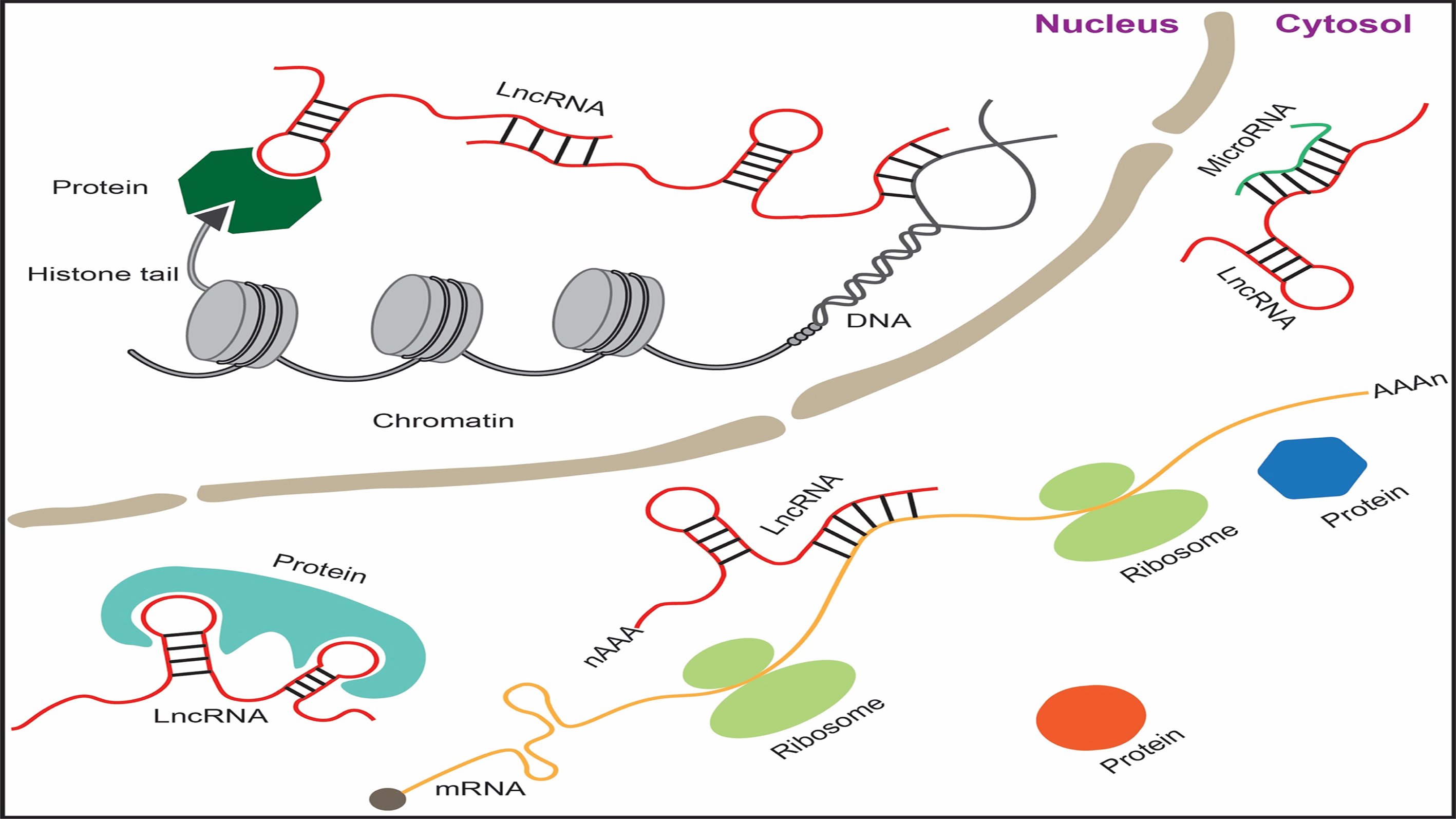
Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches
Xingyang Qian*, Jieyu Zhao*, Pui Yan Yeung*, Qiangfeng Cliff Zhang# and Chun Kit Kwok#

Advances and challenges towards the study of RNA-RNA interactions in a transcriptome-wide scale
Jing Gong*, Yanyan Ju*, Di Shao and Qiangfeng Cliff Zhang

Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate
Zhongmin Liu*, Jia Wang*, Hang Cheng*, Xin Ke*, Lei Sun, Qiangfeng Cliff Zhang, and Hong-Wei Wang

PARIS: Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution
Zhipeng Lu, Jing Gong, and Qiangfeng Cliff Zhang

RISE: a database of RNA interactome from sequencing experiments
Jing Gong*, Di Shao*, Kui Xu, Zhipeng Lu, Zhi John Lu, Yucheng T. Yang#, and Qiangfeng Cliff Zhang#,
RNA Regulations and Functions Decoded by Transcriptome-wide RNA Structure Probing
Meiling Piao, Lei Sun, Qiangfeng Cliff Zhang#

A transfer-RNA-derived small RNA regulates ribosome biogenesis
Hak Kyun Kim, Gabriele Fuchs, Shengchun Wang, Wei Wei, Yue Zhang, Hyesuk Park, Biswajoy Roy-Chaudhuri, Pan Li, Jianpeng Xu, Kirk Chu, Feijie Zhang, Mei-Sze Chua, Samuel So, Qiangfeng Cliff Zhang, Peter Sarnow, Mark A. Kay

RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer,
Li Fu*, Yan-Ru Qin*, Xiao-Yan Ming*, Xian-Bo Zuo*, Yu-Wen Diao*, Li-Yi Zhang, Jiaoyu Ai, Bei-Lei LiuTu-Xiong Huang, Ting-Ting Cao, Bin-Bin Tan, Di Xiang, Chui-Mian Zeng, Jing Gong, Qiangfeng Zhang, Sui-Sui Dong, Juan Chen, Haibo Liu, Jian-Lin Wu, Robert Z. Qi, Dan Xie, Li-Dong Wang, and Xin-Yuan Guand
Proceedings of the National Academy of Sciences 114 (23), E4631-E4640(PDF)
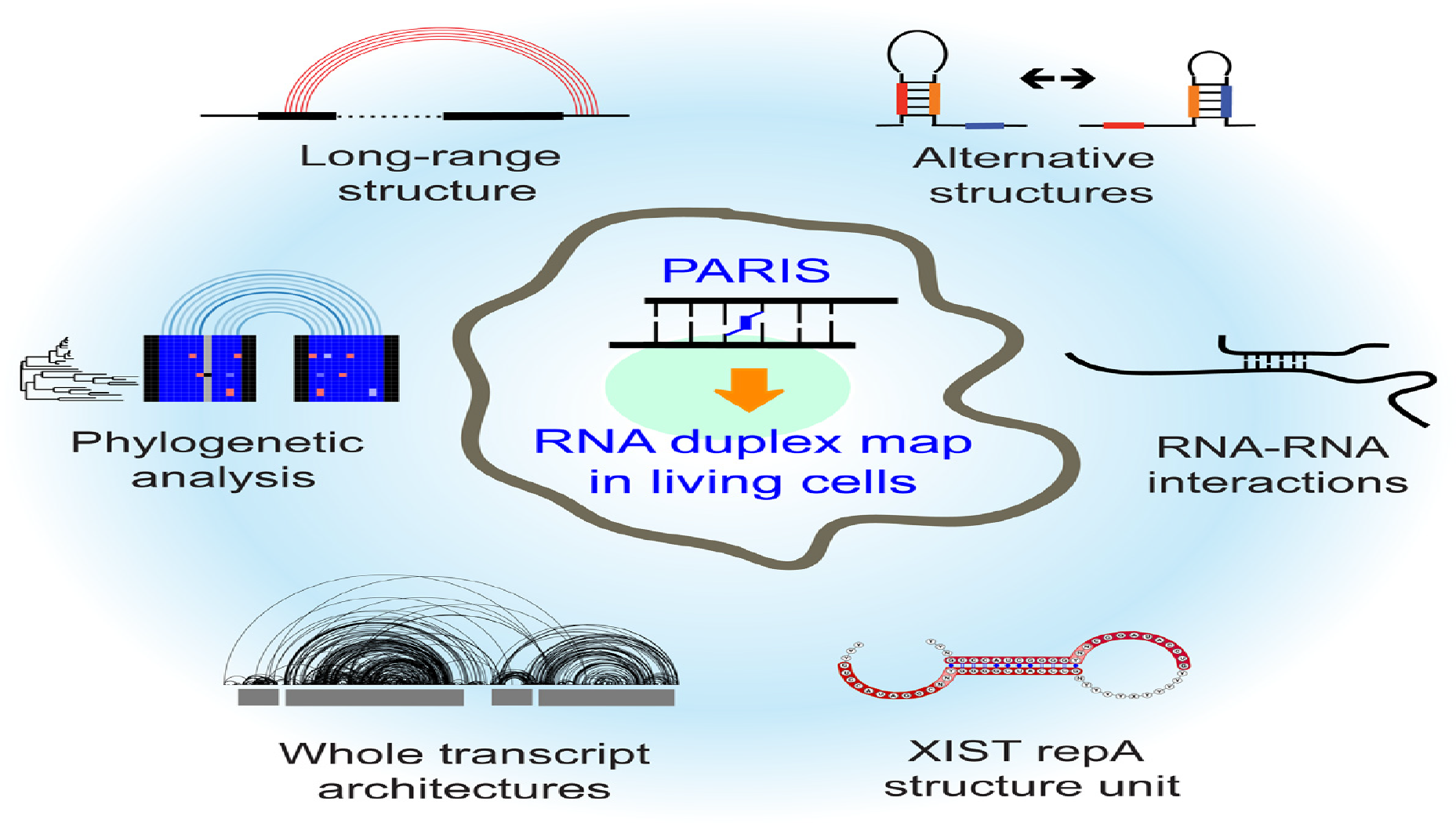
RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure
Zhipeng Lu*, Qiangfeng Cliff Zhang*, Byron Lee, Ryan A. Flynn, Martin A. Smith, James T. Robinson, Chen Davidovich, Anne R. Gooding, Karen J. Goodrich, John S. Mattick, Jill P. Mesirov, Thomas R. Cech and Howard Y. Chang

Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE
R.A. Flynn*, Q.C. Zhang*, R.C. Spitale, B. Lee, M.R. Mumbach, H.Y. Chang

Rapid evolutionary turnover underlies conserved lncRNA-genome interactions
J.J. Quinn*, Q.C. Zhang*, P. Georgiev, I.A. Ilik, A. Akhtar, H.Y. Chang

Structural imprints in vivo decode RNA regulatory mechanisms
R.C. Spitale*, R.A. Flynn*, Q.C. Zhang*, P. Crisalli, B. Lee, J.W. Jung, H.Y. Kuchelmeister, P.J. Batista, E.A. Torre, E.T. Kool, H.Y. Chang

Systematic discovery of Xist RNA binding proteins
C. Chu, Q.C. Zhang, S.T. da Rocha, R.A. Flynn, M. Bharadwaj, J.M. Calabrese, T. Magnuson, E. Heard, H.Y. Chang

Extensive variation in chromatin states across humans
M. Kasowski*, S. Kyriazopoulou-Panagiotopoulou*, F. Grubert*, J.B. Zaugg*, A. Kundaje*, Y. Liu, A.P. Boyle, Q.C. Zhang, F. Zakharia, D.V. Spacek, J. Li, D. Xie, A. Olarerin-George, L.M. Steinmetz, J.B. Hogenesch, M. Kellis, S. Batzoglou, M. Snyder

PrePPI: a structure-informed database of protein-protein interactions
Q.C. Zhang, D. Petrey, J.I. Garzon, L. Deng, B. Honig

Structure-based prediction of protein-protein interactions on a genome-wide scale
Q.C. Zhang*, D. Petrey*, L. Deng, L. Qiang, Y. Shi, C.A. Thu, B. Bisikirska, C. Lefebvre, D. Accili, T. Hunter, T. Maniatis, A. Califano, B. Honig

PredUs: a web server for predicting protein interfaces using structural neighbors
Q.C. Zhang, L. Deng, M. Fisher, J. Guan, B. Honig, D. Petrey

Protein interface conservation across structure space
Q.C. Zhang, D. Petrey, R. Norel, B.H. Honig
Proceedings of the National Academy of Sciences 107 (24), 10896-10901(PDF)